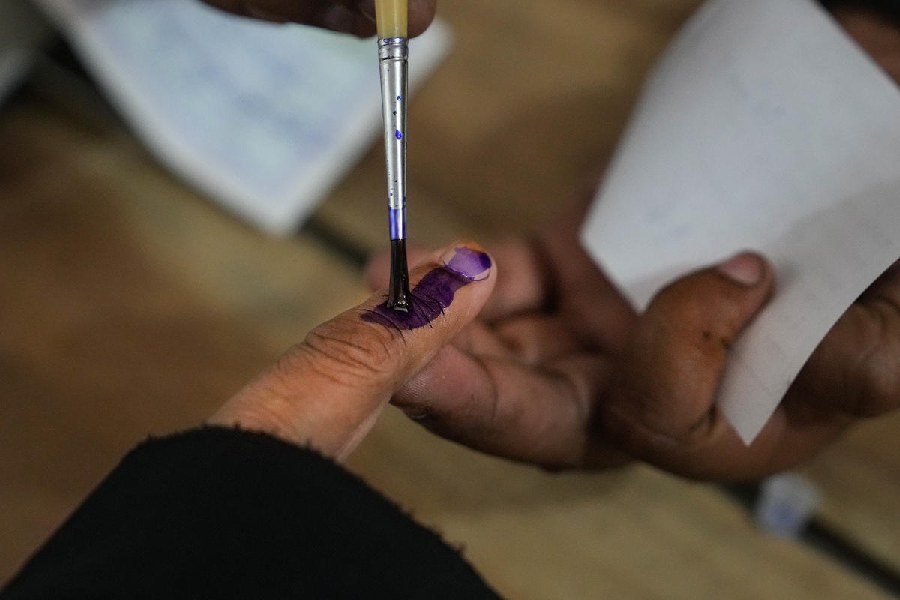
Sunday, 28 April 2024
Sunday, 28 April 2024
 Sunday, 28 April 2024
Sunday, 28 April 2024













Trina’s summer sneak out in Pattaya is a complete funventure
‘The Boy and The Heron’ is coming to Indian cinemas!
Surprise your guests by serving Bougainvillea Cooler this summer
Mango’s Barcelona-inspired Summer collection is a must in your wardrobe
‘Nivea Sun’, your sun-protection partner, is now on Nykaa
As politicians send riot police to try to smother a new protest movement, we’d do well to keep in mind why we’ve forgotten the ugliest aspects of the Vietnam protests
LYDIA POLGREEN

The scapegoating of Sri Lankan Tamils has broadly followed similar patterns as in India, even if the axis of persecution took place more on ethnic/linguistic rather than religious lines
ASIM ALI
The truths we express through our art are selected after careful consideration. Only those that skim the surface or keep our deep-seated beliefs intact find artistic expression
T.M. KRISHNA
Bengali does not have one ‘single’ word for them, although they are variously described as unmarried, widowed, separated or divorced. Nor does Hindi. Not that they need new descriptions
CHANDRIMA S. BHATTACHARYA
The extraordinary events in Rushdie's life, set off by the 1989 fatwa against him that resulted with the attempt on his life, can be made credible only by discarding logic as magic realism does
THE EDITORIAL BOARD































The Nag Ashwin Reddy directorial also features Amitabh Bachchan, Deepika Padukone, Disha Patani and Kamal Haasan in important roles